Text Mining for IDMP – Kill or Cure?
By Dieter Schlaps for IDMP1-News September 2018
-
Brief View into the History of Text Mining
The use of Text Mining for IDMP is based on historic experience. Artificial Intelligence methods to aid decision making in Medicine started probably with the Mycin expert system in the early 70’s, that used rules and heuristic reasoning[1] to identify bacteria causing severe infections, such as bacteremia and meningitis, and to recommend antibiotics, with the dosage adjusted for patient’s body weight. The name derived from the antibiotics themselves, as many antibiotics have the suffix “-mycin”. The Mycin system was also used for the diagnosis of blood clotting diseases. MYCIN was developed over five or six years in the early 1970s at Stanford University. It was written in LISP[2] as the doctoral dissertation of Edward Shortliffe under the direction of Bruce G. Buchanan, Stanley N. Cohen and others [Wikipedia].
Although Mycin was never used in medicine to assist in the diagnostic decision making it nevertheless had a great impact on the development of expert systems in various areas of decision making.
The information that Mycin needed to apply its decision making rules were obtained through very simple question-answer dialogues that were mostly of the yes/no type, therefore no more complex text analysis virtues were necessary…
Nevertheless, the analysis of text started even earlier, mostly using LISP, as it was the main programming language for Artificial Intelligence systems from the mid-1950’s on. But it took much longer, i.e. more than 3 decades until the first systems appeared that were able to tackle full-text: This indicates already the high level of complexity that is involved, when texts are to be analysed…
Full-text databases (e.g. Tieto TRIP[3]) became available that were database systems and search engines at the same time. Depending on how they are used, they can be said to be a database system with an integrated search engine, or a search engine with database features. Full-text databases do not use SQL as a query language and are structured quite differently from relational databases, as they index documents by their content, not by metadata about the documents.
Until the mid-nineties such systems were frequently to be found at the intelligence/information agencies that analysed and routed the reports of their “reporters, field-workers, spies…” to the relevant persons in the agencies, and at patent offices that analysed patent ideas, classified them and routed them to the patent examiners, etc.
With full-text databases it was already possible to search for keywords in large collections of text documents at a relatively high speed, and to improve the precision[4] and recall[5] of the searches by using synonyms, stop-words[6] and stemming[7].
With the arrival of the Internet, the text mining research finally paid off for the public, too: AltaVista was a Web search engine established in 1995. It became one of the most-used early search engines, but lost ground to Google and was purchased by Yahoo! in 2003, which retained the brand, but based all AltaVista searches on its own search engine.
As a summary, we define “text mining” or “knowledge discovery from text” as the machine supported analysis of text. It uses techniques from information retrieval, information extraction as well as natural language processing and connects them with the algorithms and methods of knowledge discovery in databases, data mining, machine learning and statistics.
The main uses of text mining are identified as follows:
- Text Encoding
- Text Classification and Clustering
- Information Extraction
- Topic Detection and Tracking
- Text Summarization
Today, several general purpose as well as specialized text mining solutions are available on the market that promise to turn “piles of free text and structured data” into “actionable knowledge” and “valuable business insights” (Megaputer).
-
Text Mining for IDMP Applications in Drug Information Data Management
In the Pharmaceutical Industry, there are a variety of potential use cases for Text Mining for IDMP. Here are just three examples:
- Sentiment Analysis in Social Networks:
The rapid increase in data and activities on social media creates a need for mining such data to get valuable insights about products and product use.
For pharmaceutical companies who are interested to collect customer experiences at first hand, sentiment analysis may be very useful, not only for Marketing, as it also may offer valuable information for market surveillance and drug safety departments. - Satisfy Health Authorities Thirst for more Pharmaceutical Product Data:
IDMP[8] [3], the new ISO standard for pharmaceutical product data requires that pharmaceutical companies collect, encode and submit an unprecedented amount of pharmaceutical product data to the Health Authorities. As the IDMP data record is regarded as the structured representation of the documents in the product dossier (and will replace them increasingly in the product submissions in the years to come…), it is intriguing to extract the data to be submitted directly from the documents via text mining and use the text encoding and data extraction functions to collect the data items.
We will cover that topic more in depth in the next chapter. - Signal Event Detection in Drug Safety/ Pharmacovigilance:
Early identification of the hazards associated with drugs is the main goal of those involved in pharmacovigilance ‘Signal detection’, ‘signal generation’ or ‘signalling’ refers to a process that aims to find, as soon as possible, any indication of an unexpected drug safety problem which may be either new ADRs or a change of the frequency of ADRs that are already known to be associated with the drugs involved. The results of this surveillance exercise tend to arouse suspicions and should always be followed up by in-depth investigations.
The sources for signal detection are publications, scientific reports, but also statements from health care professionals and/or patients that are made public through social networks other internet sources, or anonymous electronic health records.
-
A Look at Text Mining for IDMP Approach to Support the Analysis of SmPC Documents
To extract the IDMP data that is expected for the first “iteration” of IDMP in Europe, especially, indication, drug manufacturer and composition data, a Text Mining for IDMP approach already seems to be attractive as – depending on the portfolio of the respective company – several ten thousands of registered products may be in the scope for IDMP submissions.
Indications, packaging, composition and further product information may be found in the Summary of Product Characteristics (“S(m)PC”), whereas the details about the involved manufacturers and the manufactured composition are found in the dossier documents (module 3).
Since the S(m)PC document accompanies a product’s registration in a country or region, it is generally written in several languages, and it may differ content wise (slightly) from country to country. The dossier, however, is the same for all products of the same product family, and for all their registrations. It normally is written in one language only (e.g. English). Therefore, the number of module 3 documents is significantly lower than the number of S(m)PCs, several hundreds in contrast to several ten thousands…
The following table summarizes the challenges and experiences with Text Mining for IDMP software for the different document types:
| Module 3.2.P.1 Composition documents | Module 3.2.P.3.1 Manufacturing documents | SmPC documents | |
|
Text mining functions needed: – Text Encoding – Text Classification and Clustering – Information Extraction – Topic Detection and Tracking – Text Summarization |
Coding of substances according G-SRS/ EU-SMS not needed Moderate Not needed |
Coding of manufacturers and manufacturer steps acc. EU OMS and RMS not needed Easy Not needed |
Extensive and additional coding needed, EU-RMS In the indication section, free text will have to be classified according to EU-RMS Can be very difficult (see below) Not needed |
| Information to be extracted |
– Active substance, excipients with their respective roles. Active substances with strengths and eventually reference substances, reference strengths. – Dose Forms |
– Manufacturer company name – Manufacturer address data – Manufacturing roles (e.g. “Secondary packaging”) |
– Product names and name components – Dose forms – Indications with intended effects, co-morbidity and age/population specifics – Paediatric use – Administration route – Unit of presentation – ATC Code – Shelf life and storage – MAH Name and address – Marketing Authorization – Packaging Description – Package containers and packaged items with quantities |
| Challenges |
– Sometimes the list of ingredients is described in a tabular way, sometimes lists are used. – When a product has different strengths, the compositions may vary. |
– The manufacturing role is often not clearly described and easily mappable to a controlled vocabulary |
– In one SmPC there may be more than one Medicinal Product (e.g. characterized by different strengths) described – In one physical document there may be one or more S(m)PCs (with one or more products) – Some of the IDMP fields are the same for all products described in SmPC (Indications, ATC code, authorisation information), others may or may not depend on the individual product (strength) – To extract information on intended effects, age and/or race characteristics may require a certain degree of text understanding. |
| Results | In general, today’s text mining products that support the IDMP data extraction and coding of product documentation are quite well able to extract the substance and/or manufacturer data from the corresponding dossier documents. | The situation is quite different for SmPCs. The results depend very much on the complexity of the SmPC’s and the ability of the respective software vendor to map the extracted data correctly to the IDMP data model (see figure below). To our knowledge, there is no vendor on the market that is able to handle SmPCs adequately and reliably from scratch.Significant tuning, configuration and testing are necessary to come up with a solution setup that is able to achieve acceptable levels of quality. |
|
Table 1: Experiences with Text Mining in IDMP Product Data Extraction (Scope: EU’s IDMP Iteration 1)
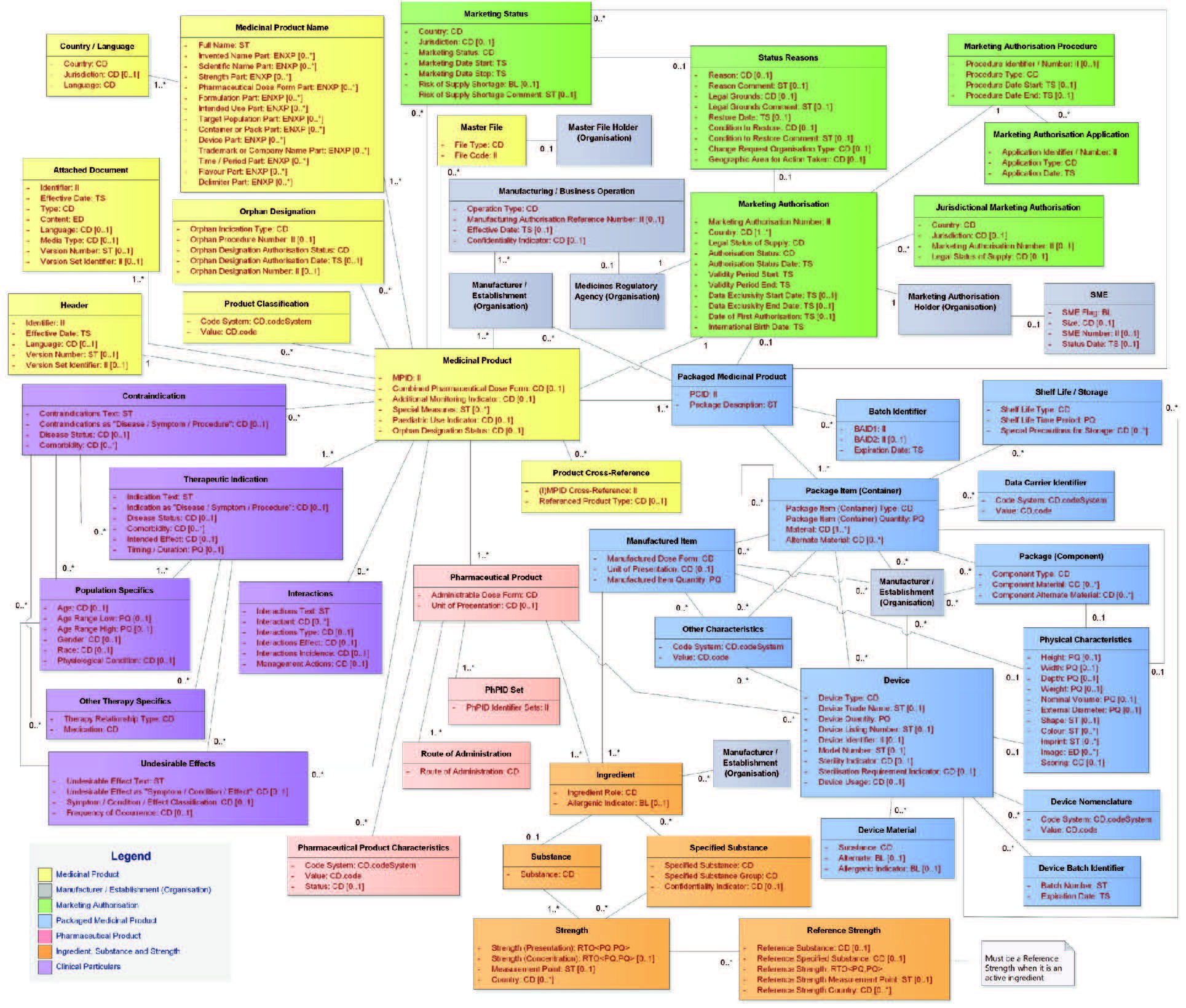
Figure 1: The (full) IDMP Data Model (Version 2)
-
Conclusions and Outlook to Text Mining for IDMP
Although the foundations of Text Mining for IDMP have been created already more than 60 years ago, and – in the meantime – powerful Text Mining for IDMP solutions are available the market place, the extraction of IDMP data from the original documents has still its challenges, which are often related to the complexity of the SmPC structures. In order to assign the data extracted from the SmPCs to the right data fields of the IDMP data structure (see previous figure), the software must be capable of understanding the structure of the SmPC and how it maps to the IDMP data structure. This not only concerns the different strengths described in the SmPC that define different products, but also e.g. the description of the packaging, with its different layers, and eventually several pharmaceutical products and/or devices being available in the same package.
In contrast to this, the extraction of text and the coding in e.g. MedDRA, G-SRS seem to be less difficult.
A suggestion to the vendors of text mining therefore is to analyse and adopt the IDMP data model more closely and to understand how it relates to the structure of SmPC documents.
Marketing authorization holders should consider the use of better structured templates, especially for SmPCs, that allow better automatic analysis, e.g. by clearly relating data field values to either an individual product or to all products. At a later stage, should the company decide to introduce content-based authoring, this would be necessary anyway…
References:
[1] Andreas Hotho, Andreas Nürnberger, Gerhard Paaß: A Brief Survey of Text Mining. (PDF) In: Zeitschrift für Computerlinguistik und Sprachtechnologie. 20, Nr. 1, 2005. Accessed on August 10, 2018. Link: https://www.kde.cs.uni-kassel.de/hotho/pub/2005/hotho05TextMining.pdf [2] Contrasting Relational Databases and Full-Text Search Engines. Last Updated Jan 2009. By: Miles Kehoe, New Idea Engineering, Inc., Volume 2 – Issue 1 – June 2004.Link: http://www.ideaeng.com/database-full-text-search-0201 [3] ISO Standards and Implementation Guidelines:
- ISO 11615, Health informatics — Identification of medicinal products — Data elements and structures for the unique identification and exchange of regulated medicinal product information;
- ISO/TS 20443, Health informatics — Identification of Medicinal Products — Implementation Guide for EN ISO 11615 Data Elements, Structures and Message Specifications for Unique Identification and Exchange of Regulated Medicinal Product Information;
- ISO 11616, Health informatics — Identification of medicinal products — Data elements and structures for the unique identification and exchange of regulated pharmaceutical product information;
- ISO/TS 20451, Health informatics – Identification of Medicinal Products – Implementation Guide for EN ISO 11616 Data elements and structures for the unique identification and exchange of regulated pharmaceutical product information;
- ISO 11238, Health informatics — Identification of medicinal products — Data elements and structures for the unique identification and exchange of regulated information on substances;
- ISO/TS 19844, Health informatics — Identification of Medicinal Products – Implementation Guide of EN ISO 11238 Data Elements and Structures for the Unique Identification and Exchange of Regulated information on Substances;
- ISO 11239, Health informatics — Identification of medicinal products — Data elements and structures for the unique identification and exchange of regulated information on pharmaceutical dose forms, units of presentation, routes of administration and packaging;
- ISO/TS 20440, Health informatics — Identification of Medicinal Products — Implementation Guide for EN ISO 11239 Data elements and structures for the unique identification and exchange of regulated information on pharmaceutical dose forms, units of presentation, routes of administration and packaging;
- ISO 11240, Health informatics — Identification of medicinal products — Data elements and structures for the unique identification and exchange of units of measurement.
[1] A heuristic technique (/hjʊəˈrɪstɪk/; Ancient Greek: εὑρίσκω, “find” or “discover”), often called simply a heuristic, is any approach to problem solving, learning, or discovery that employs a practical method, not guaranteed to be optimal, perfect, logical, or rational, but instead sufficient for reaching an immediate goal. Where finding an optimal solution is impossible or impractical, heuristic methods can be used to speed up the process of finding a satisfactory solution. Heuristics can be mental shortcuts that ease the cognitive load of making a decision. Examples that employ heuristics include using a rule of thumb, an educated guess, an intuitive judgment, a guesstimate, stereotyping, profiling, or common sense. (Wikipedia 2018)
[2] LISP: List Processing Language: Lisp was invented by John McCarthy in 1958 while he was at the Massachusetts Institute of Technology (MIT). McCarthy published its design in a paper in Communications of the ACM in 1960, entitled “Recursive Functions of Symbolic Expressions and Their Computation by Machine, Part I”
[3] TRIP was originally developed in Sweden by Paralog AB and has over 30 years of history and many installations. Since 1999 TRIP has been wholly owned and developed by Tieto.
[4] In the field of information retrieval, precision is the fraction of retrieved documents that are relevant to the query. For example, for a text search on a set of documents, precision is the number of correct results divided by the number of all returned results.
[5] In information retrieval, recall is the fraction of the relevant documents that are successfully retrieved.
For example, for a text search on a set of documents, recall is the number of correct results divided by the number of results that should have been returned.
[6] In computing, stop words are words which are filtered out before or after processing of natural language data (text). Though “stop words” usually refers to the most common words in a language, there is no single universal list of stop words used by all natural language processing tools, and indeed not all tools even use such a list. Some tools specifically avoid removing these stop words to support phrase search.
[7] In linguistic morphology and information retrieval, stemming is the process of reducing inflected (or sometimes derived) words to their word stem, base or root form—generally a written word form.
[8] Identification of Medicinal Products, see [3]

